TADKit - Interactive Anomaly Detection demonstrator
Loading use case
Where you import your data. We simply generate some synthetic timeseries. We either use the synchronous format where each timeseries is a column with label as a column name, or the asynchronous format with the two columns (“sensor”, “data”) where “sensor” has the timeseries labels and “data” their corresponding values.
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
import matplotlib.pyplot as plt; plt.rcParams['figure.figsize'] = [11, 5]
import numpy as np
import pandas as pd
from tadkit.utils.synthetic_ornstein_uhlenbeck import synthetise_ornstein_uhlenbeck_data
X, y = synthetise_ornstein_uhlenbeck_data(n_rows=10000, n_cols_x=20)
display(X)
async_X = X.melt(value_vars=X.columns.tolist(), ignore_index=False).rename(
columns={"variable": "sensor", "value": "data"}
)
display(async_X)
pd.concat([X, y], axis=1).plot()
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 | X13 | X14 | X15 | X16 | X17 | X18 | X19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021-01-01 00:00:00 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 2021-01-01 01:00:00 | 0.001661 | 0.007820 | 0.008523 | -0.007071 | -0.009317 | 0.008867 | -0.002218 | 0.003817 | -0.007726 | 0.008630 | -0.002813 | -0.009319 | -0.004985 | 0.007643 | 0.001915 | -0.006188 | 0.016657 | 0.017382 | 0.011822 | 0.011194 |
| 2021-01-01 02:00:00 | 0.008823 | -0.010364 | 0.019886 | -0.007703 | -0.007919 | 0.002534 | -0.008656 | 0.010696 | -0.018860 | 0.009704 | -0.013080 | -0.013086 | 0.008871 | 0.004319 | -0.010231 | -0.009694 | 0.017622 | 0.026136 | 0.014752 | 0.005832 |
| 2021-01-01 03:00:00 | -0.002155 | -0.019065 | 0.011051 | -0.003709 | -0.007369 | -0.005963 | 0.001210 | 0.013652 | -0.001756 | 0.005926 | -0.015816 | -0.024475 | 0.004826 | -0.004678 | -0.014786 | -0.007505 | 0.024207 | 0.014618 | 0.013044 | -0.004295 |
| 2021-01-01 04:00:00 | 0.013071 | -0.016399 | -0.006838 | 0.002436 | 0.000271 | -0.016800 | 0.013201 | -0.000847 | 0.009786 | 0.001089 | -0.022053 | -0.047641 | -0.001618 | -0.011110 | -0.010734 | -0.026655 | 0.029129 | 0.024849 | 0.024760 | -0.006651 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2022-02-21 11:00:00 | 0.480157 | 1.036571 | 0.079168 | -1.287169 | 2.634510 | -0.210555 | -0.207754 | 2.790915 | 1.994599 | 1.263292 | -0.291272 | 0.304258 | 0.506799 | 0.362485 | 2.633189 | 0.370203 | -1.510844 | 1.593467 | -1.383251 | 0.287831 |
| 2022-02-21 12:00:00 | 0.472759 | 1.028854 | 0.078058 | -1.292116 | 2.634895 | -0.217651 | -0.222453 | 2.803378 | 2.005869 | 1.255607 | -0.292545 | 0.306667 | 0.504882 | 0.369449 | 2.632624 | 0.357541 | -1.516517 | 1.592511 | -1.402227 | 0.300942 |
| 2022-02-21 13:00:00 | 0.484448 | 1.035954 | 0.077118 | -1.286552 | 2.640465 | -0.218561 | -0.227952 | 2.794754 | 2.003116 | 1.252523 | -0.297669 | 0.301481 | 0.492823 | 0.380544 | 2.612742 | 0.354779 | -1.513238 | 1.600206 | -1.388866 | 0.297647 |
| 2022-02-21 14:00:00 | 0.478087 | 1.037739 | 0.066010 | -1.273569 | 2.644860 | -0.223948 | -0.225738 | 2.787982 | 2.008781 | 1.265143 | -0.302747 | 0.285736 | 0.491451 | 0.382196 | 2.613877 | 0.356197 | -1.517774 | 1.585939 | -1.385370 | 0.288772 |
| 2022-02-21 15:00:00 | 0.474328 | 1.052743 | 0.068460 | -1.276385 | 2.637026 | -0.225501 | -0.232017 | 2.779689 | 2.006385 | 1.254136 | -0.304361 | 0.281531 | 0.502489 | 0.386042 | 2.608148 | 0.352264 | -1.510109 | 1.595649 | -1.392864 | 0.284664 |
10000 rows × 20 columns
| sensor | data | |
|---|---|---|
| 2021-01-01 00:00:00 | X0 | 0.000000 |
| 2021-01-01 01:00:00 | X0 | 0.001661 |
| 2021-01-01 02:00:00 | X0 | 0.008823 |
| 2021-01-01 03:00:00 | X0 | -0.002155 |
| 2021-01-01 04:00:00 | X0 | 0.013071 |
| ... | ... | ... |
| 2022-02-21 11:00:00 | X19 | 0.287831 |
| 2022-02-21 12:00:00 | X19 | 0.300942 |
| 2022-02-21 13:00:00 | X19 | 0.297647 |
| 2022-02-21 14:00:00 | X19 | 0.288772 |
| 2022-02-21 15:00:00 | X19 | 0.284664 |
200000 rows × 2 columns
<Axes: >
Formalizer: given a use case, define how to formalize the dataset for ML
Select a Formalizer: tadkit-provided object adapted to the use case type (synchronous or asynchronous) you are set on learning.
from tadkit.catalog.formalizers import PandasFormalizer
from tadkit.utils.widget import select_formalizer
formalizers = {
"synchronous_formalizer": PandasFormalizer(
data_df=X,
dataframe_type="synchronous",
),
"asynchronous_formalizer": PandasFormalizer(
data_df=async_X,
dataframe_type="asynchronous",
)
}
for formalizer_name, formalizer in formalizers.items():
print(f"{formalizer_name} has {formalizer.available_properties=}")
formalizer_selector = select_formalizer(formalizers)
formalizer_selector
[TADKit-Catalog]
Class learner_name='cnndrad' is registered in TADKit.
cnndrad returns err=ModuleNotFoundError("No module named 'cnndrad'").
Class learner_name='sbad' is registered in TADKit.
sbad returns err=ModuleNotFoundError("No module named 'sbad_fnn'").
Class learner_name='kcpd' is registered in TADKit.
kcpd returns err=ModuleNotFoundError("No module named 'kcpdi'").
Class learner_name='tdaad' is registered in TADKit.
tdaad returns err=ModuleNotFoundError("No module named 'tdaad'").
Class learner_name='isolation-forest' is registered in TADKit.
isolation-forest is operational in this environment.
isolation-forest is implicit child of TADLearner.
Class learner_name='kernel-density' is registered in TADKit.
kernel-density is operational in this environment.
kernel-density is implicit child of TADLearner.
Class learner_name='scaled-kernel-density' is registered in TADKit.
scaled-kernel-density is operational in this environment.
scaled-kernel-density is implicit child of TADLearner.
synchronous_formalizer has formalizer.available_properties=['pandas', 'fixed_time_step']
asynchronous_formalizer has formalizer.available_properties=['pandas', 'fixed_time_step']
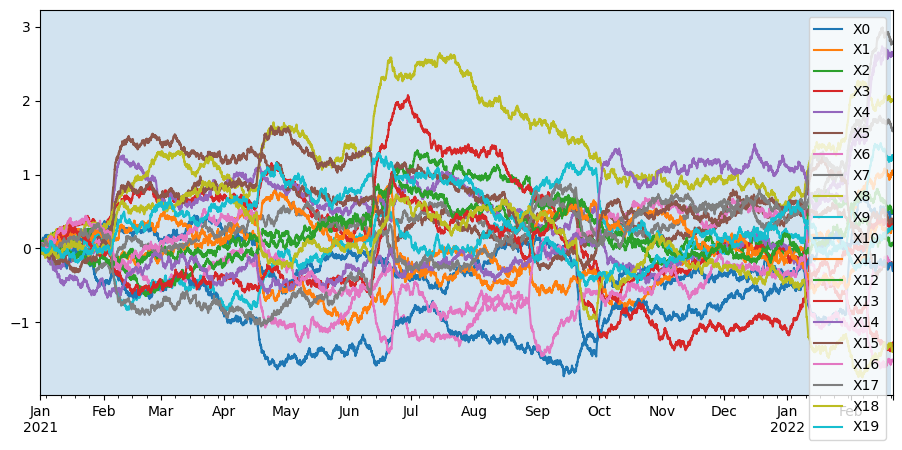
This allows to select a training interval, a resampling resolution and the set of target sensors for learning.
from tadkit.utils.widget import query_widget_selection
formalizer_name = formalizer_selector.value
query_dict = query_widget_selection(formalizer_name, formalizers[formalizer_name].query_description)
'synchronous_formalizer'
query_translated = {key: widget.value for key, widget in query_dict.items()}
X_train = formalizers[formalizer_name].formalize(**query_translated)
query_translated.pop("target_period")
X_test = formalizers[formalizer_name].formalize(**query_translated)
ax = X_test.plot()
ax.axvspan(X_train.index[0], X_train.index[-1], alpha=0.2)
plt.show()
The dataset formalizing is done and ready for learning.
TADLearner: detect anomalies
from tadkit.catalog.learners import installed_learner_classes
Select a set of learners that are compatible with the formalizing.
from tadkit.utils.widget import select_matching_available_learners
matching_available_learners = select_matching_available_learners(formalizers[formalizer_name], installed_learner_classes)
Finally, set the parameters of the chosen learners.
from tadkit.utils.widget import parameter_widget_selection
learners_params = {
learner_name: parameter_widget_selection(
tad_object_name=learner_name,
params_description=installed_learner_classes[learner_name].params_description,
) for learner_name in matching_available_learners.value
}
>>> Parameters for learner isolation-forest
>>> Parameters for learner kernel-density
>>> Parameters for learner scaled-kernel-density
learners = {
learner_class_name:
installed_learner_classes.get(learner_class_name)(
**{param: selection.value for (param, selection) in params_dict.items()}
)
for learner_class_name, params_dict in learners_params.items()
}
learners
{'isolation-forest': IsolationForest(n_estimators=10),
'kernel-density': KernelDensity(kernel='epanechnikov'),
'scaled-kernel-density': ScaledKernelDensityLearner(scaling='quantile_normal')}
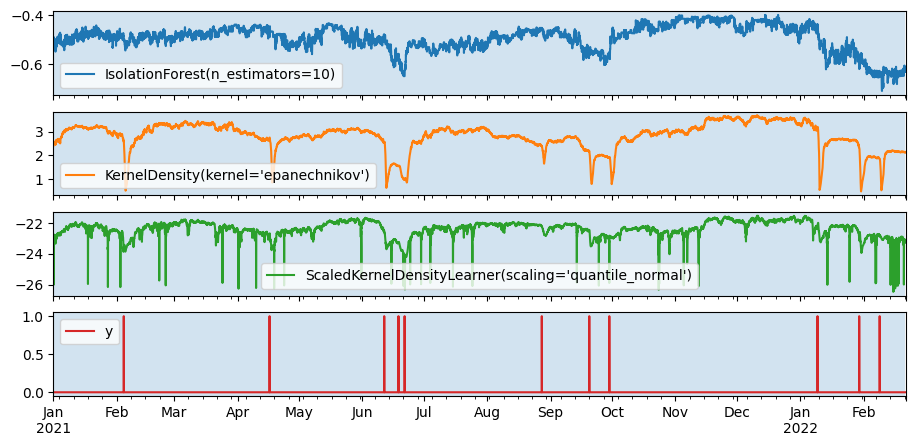
Fit on train query, score on the entire series.
# Fit loop:
anomalies = pd.DataFrame(index=X_test.index)
for learner_name, learner_object in learners.items():
print(f"Fitting {learner_name}")
learner_object.fit(X_train)
# Score loop:
for learner_name, fitted_learner_object in learners.items():
print(f"Scoring with {learner_name}")
anom_score = fitted_learner_object.score_samples(X_test)
anomalies[str(fitted_learner_object)] = anom_score
axes = pd.concat([anomalies, y], axis=1).plot(subplots=True)
[ax.axvspan(X_train.index[0], X_train.index[-1], alpha=0.2) for ax in axes]
plt.show()
Fitting isolation-forest
Fitting kernel-density
Fitting scaled-kernel-density
Scoring with isolation-forest
Scoring with kernel-density
Scoring with scaled-kernel-density
from plotly.offline import init_notebook_mode, iplot
from plotly.graph_objs import *
init_notebook_mode(connected=True) # initiate notebook for offline plot
pd.options.plotting.backend = "plotly"
fig = pd.concat([anomalies.apply(lambda x: (x - x.min()) / (x.max() - x.min())), y], axis=1).plot()
fig.update_layout(
legend=dict(
orientation="v",
entrywidth=100,
yanchor="bottom",
y=1.02,
xanchor="right",
x=1
),
width=1000,
height=600,
)
fig.show()
pd.options.plotting.backend = "matplotlib"