import sys
from pprint import pprint
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [11, 5]
from sklearn.ensemble import IsolationForest
from sklearn.preprocessing import StandardScaler
PROJECT_DIR = '..'
if PROJECT_DIR not in sys.path:
sys.path.insert(0, PROJECT_DIR)
from tadkit.base import TADLearner
from tadkit.catalog.formalizers import PandasFormalizer
Synthetic data simulation
from tadkit.utils.synthetic_ornstein_uhlenbeck import synthetise_ornstein_uhlenbeck_data
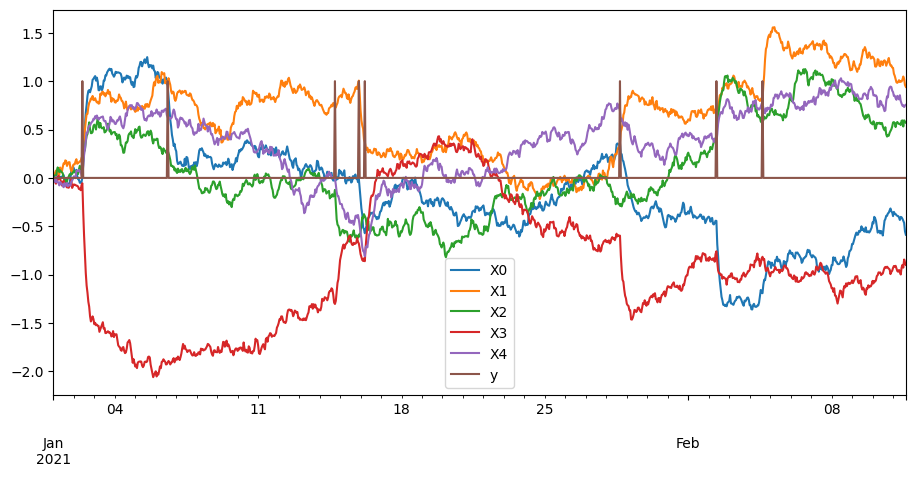
X, y = synthetise_ornstein_uhlenbeck_data(n_rows=1000, n_cols_x=5)
pd.concat((X, y), axis=1).plot()
<Axes: >
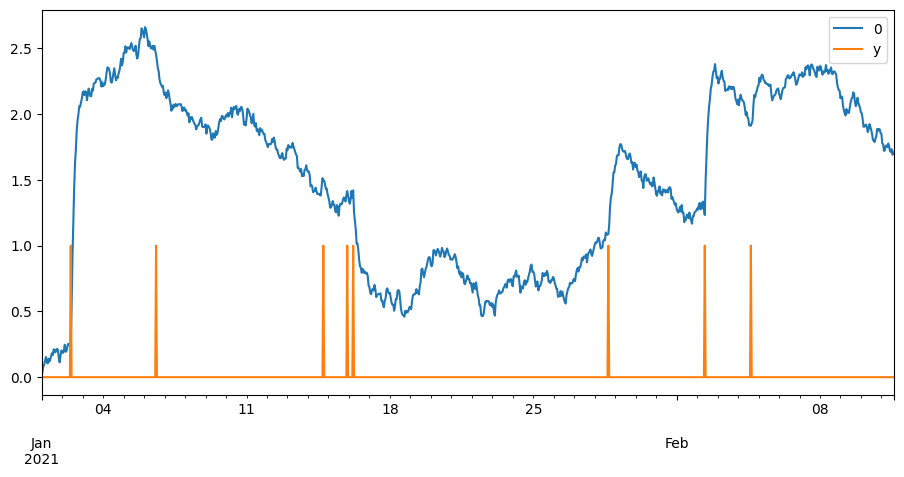
pd.concat([((X ** 2).sum(axis=1) ** .5), y], axis=1).plot()
<Axes: >
Formalizer
formalizer = PandasFormalizer(
data_df=X,
dataframe_type="synchronous",
)
formalizer.available_properties
['pandas', 'fixed_time_step']
pprint(formalizer.query_description)
{'resampling': {'default': False,
'description': 'Resampling of the target query',
'family': 'bool'},
'resampling_resolution': {'default': 120,
'description': 'If resampling, resampling '
'resolution in seconds.',
'family': 'time',
'start': 60,
'stop': 3600},
'target_period': {'default': (Timestamp('2021-01-01 00:00:00'),
Timestamp('2021-02-11 15:00:00')),
'description': 'Time period for your query.',
'family': 'time_interval',
'start': Timestamp('2021-01-01 00:00:00'),
'stop': Timestamp('2021-02-11 15:00:00')},
'target_space': {'default': Index(['X0', 'X1', 'X2', 'X3', 'X4'], dtype='object'),
'description': 'List of sensors used for your query.',
'family': 'space',
'set': Index(['X0', 'X1', 'X2', 'X3', 'X4'], dtype='object')}}
formalizer.formalize(data=X, columns_0=['X1', 'X2'],
time_interval=['2021-01-01 02:00:00', '2021-02-02'],
target_space=X.columns,
resampling=True,
resampling_resolution=60)
| X0 | X1 | X2 | X3 | X4 | |
|---|---|---|---|---|---|
| 2021-01-01 00:00:00 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 2021-01-01 00:01:00 | 0.000088 | 0.000412 | 0.000449 | -0.000373 | -0.000491 |
| 2021-01-01 00:02:00 | 0.000175 | 0.000824 | 0.000898 | -0.000745 | -0.000982 |
| 2021-01-01 00:03:00 | 0.000263 | 0.001236 | 0.001348 | -0.001118 | -0.001473 |
| 2021-01-01 00:04:00 | 0.000350 | 0.001649 | 0.001797 | -0.001491 | -0.001964 |
| ... | ... | ... | ... | ... | ... |
| 2021-02-11 14:56:00 | -0.584958 | 0.942160 | 0.571343 | -0.897471 | 0.755402 |
| 2021-02-11 14:57:00 | -0.585516 | 0.941845 | 0.571196 | -0.897901 | 0.755519 |
| 2021-02-11 14:58:00 | -0.586074 | 0.941530 | 0.571048 | -0.898330 | 0.755637 |
| 2021-02-11 14:59:00 | -0.586633 | 0.941214 | 0.570901 | -0.898760 | 0.755755 |
| 2021-02-11 15:00:00 | -0.587191 | 0.940899 | 0.570754 | -0.899189 | 0.755873 |
59941 rows × 5 columns
Learners
print(isinstance(IsolationForest, TADLearner))
from tadkit.utils.tadlearner_factory import tadlearner_factory
IsolationForestLearner = tadlearner_factory(IsolationForest, [], {})
print(isinstance(IsolationForestLearner, TADLearner))
False
True
from tadkit.utils.decomposable_tadlearner import decomposable_tadlearner_factory
StandardForestLearner = decomposable_tadlearner_factory(StandardScaler, IsolationForestLearner, [], {})
learner = StandardForestLearner()
learner.random_state = -1
learner.fit(X)
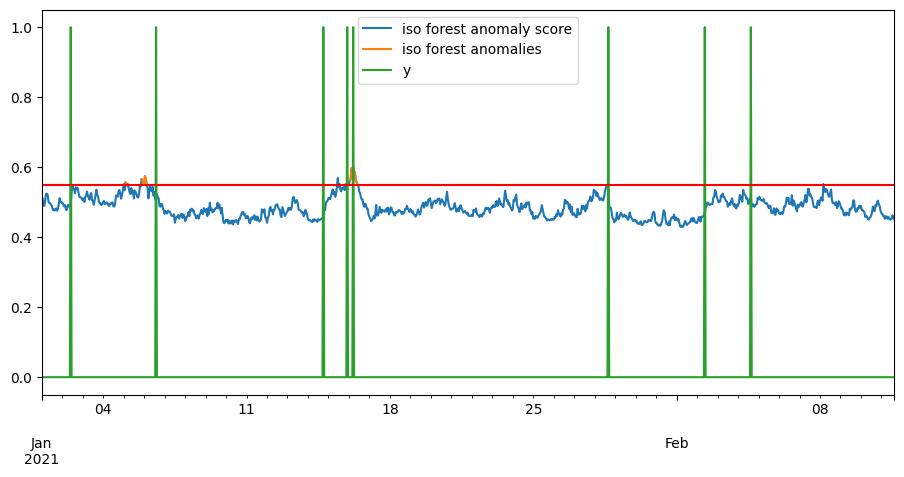
anom_score = -learner.score_samples(X)
iso_score = pd.DataFrame(anom_score, index=X.index, columns=["iso forest anomaly score"])
ceil = .55
iso_anomalies = iso_score[iso_score > ceil]
iso_anomalies.rename(columns={"iso forest anomaly score": "iso forest anomalies"}, inplace=True)
pd.concat([iso_score, iso_anomalies, y], axis=1).plot()
plt.axhline(y=ceil, color='r', linestyle='-')
learner
StandardScalerIsolationForest()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
StandardScalerIsolationForest()
class ExponantialMean:
def __init__(self, lags, sub_X=False) -> None:
self.lags = np.array(lags)
self.sub_X = sub_X
assert sum(d != 1 for d in self.lags.shape) <= 1
self.lags = self.lags.reshape(-1)
def transform(self, X):
lags_len, = self.lags.shape
index = X.index
columns = sum(
([f'{colname}_ema{i}' for i in range(lags_len)] for colname in X.columns),
start=[]
)
t = index.astype('int64') * 1e-9
X = X.values
n, d = X.shape
result = np.empty((n, d, lags_len))
result[0] = X[0, :, None]
for i in range(n-1):
weight = np.exp(-(t[i+1] - t[i]) / self.lags)[None, None]
result[i+1] = result[i] * weight + X[i+1, :, None] * (1 - weight)
if self.sub_X:
result -= X[:, :, None]
result = pd.DataFrame(
result.reshape((n, -1)),
index=index,
columns=columns
)
return result
MINUTE = 60
HOUR = 60 * MINUTE
DAY = 24 * HOUR
exponential_mean = ExponantialMean([DAY, 3* DAY], sub_X=True)
exponential_mean.transform(X)
| X0_ema0 | X0_ema1 | X1_ema0 | X1_ema1 | X2_ema0 | X2_ema1 | X3_ema0 | X3_ema1 | X4_ema0 | X4_ema1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2021-01-01 00:00:00 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 2021-01-01 01:00:00 | -0.005038 | -0.005180 | -0.023719 | -0.024387 | -0.025852 | -0.026580 | 0.021447 | 0.022051 | 0.028259 | 0.029055 |
| 2021-01-01 02:00:00 | -0.031711 | -0.032745 | -0.048762 | -0.050794 | -0.001294 | -0.002048 | 0.037732 | 0.039390 | 0.037896 | 0.039748 |
| 2021-01-01 03:00:00 | -0.015200 | -0.016647 | -0.069807 | -0.073777 | -0.007043 | -0.007985 | 0.054845 | 0.058025 | -0.014293 | -0.012868 |
| 2021-01-01 04:00:00 | -0.067253 | -0.070574 | -0.048571 | -0.053855 | -0.062409 | -0.065097 | 0.030707 | 0.034709 | 0.041473 | 0.044046 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-02-11 11:00:00 | -0.051206 | -0.211574 | 0.117038 | 0.144954 | -0.001470 | 0.090219 | -0.091695 | -0.115633 | 0.091504 | 0.073529 |
| 2021-02-11 12:00:00 | -0.028717 | -0.187681 | 0.081779 | 0.111612 | 0.058907 | 0.150990 | -0.060468 | -0.085780 | 0.081817 | 0.066394 |
| 2021-02-11 13:00:00 | -0.007902 | -0.164897 | 0.104148 | 0.136504 | -0.004352 | 0.086338 | -0.138787 | -0.167659 | 0.096796 | 0.084312 |
| 2021-02-11 14:00:00 | 0.077915 | -0.074720 | 0.158832 | 0.195215 | 0.010635 | 0.100374 | -0.104210 | -0.135619 | 0.077579 | 0.067452 |
| 2021-02-11 15:00:00 | 0.106862 | -0.040657 | 0.170498 | 0.211182 | 0.018675 | 0.107702 | -0.075242 | -0.108337 | 0.067626 | 0.059545 |
1000 rows × 10 columns
EMAForestLearner = decomposable_tadlearner_factory(ExponantialMean, IsolationForestLearner, [], {
'lags': {
'description': 'Lags typical time of exponential mean',
'family': 'preprossing',
'value_type': 'set',
'start': 0,
},
'sub_X': {
'description': 'Value substract from to the lags',
'family': 'preprossing',
'value_type': 'bool',
},
})
learner = EMAForestLearner(lags=np.geomspace(.1 * DAY, 2 * DAY, 10), sub_X=True)
learner.random_state = -1
learner.fit(X)
anom_score = -learner.score_samples(X)
ema_score = pd.DataFrame(anom_score, index=X.index, columns=["ema iso forest anomaly score"])
ceil = .5
ema_anomalies = ema_score[iso_score > ceil]
ema_anomalies.rename(columns={"ema iso forest anomaly score": "ema iso forest anomalies"}, inplace=True)
learner
ExponantialMeanIsolationForest()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
ExponantialMeanIsolationForest()
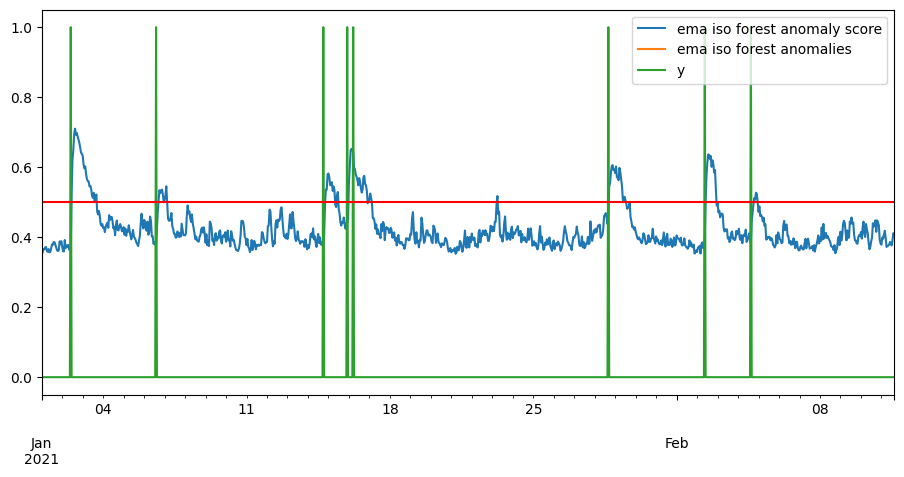
pd.concat([ema_score, ema_anomalies, y], axis=1).plot()
plt.axhline(y=ceil, color='r', linestyle='-')
<matplotlib.lines.Line2D at 0x29528e1e630>